ge03
Emily Ressler
4/19/2018
Exercise 1
library(tidyverse) #loads tidyverse package
#import data into R
monthly_temps <- read_csv("monthly_global_land_and_ocean_temperature_anomalies.csv")## Parsed with column specification:
## cols(
## YearMonth = col_integer(),
## Value = col_double()
## )library(lubridate) #loads lubridate##
## Attaching package: 'lubridate'## The following object is masked from 'package:base':
##
## date#convert class from integer to date, day is missing
monthly_temps <- mutate(monthly_temps, date = ymd(YearMonth, truncated=1))
# year and month stored as separate variables, keep name of month
monthly_temps <- mutate(monthly_temps,
year=year(date),
month=month(date,label=TRUE)
)
#rename value --> temperature_anomaly
monthly_temps <- rename(monthly_temps, temperature_anomaly = Value)
#drop the YearMonth variable (replaced by date variable)
monthly_temps <- select(monthly_temps, -YearMonth)Exercise 2
You can also embed plots, for example:
library(tidyverse)
#import data into R
gridded_data <- read_csv("gridded_data.csv",col_names=F)## Parsed with column specification:
## cols(
## .default = col_integer()
## )## See spec(...) for full column specifications.#create variables for number of months and for monthly data
number_of_months <- nrow(gridded_data)/37
monthly_data <- vector("list",number_of_months)
library(lubridate)
#for loop
for(i in 1:number_of_months){
#extract and store temperature anomalies
monthly_data[[i]] <- gridded_data[((i-1)*37+2):(i*37),]
#get date for current month in yyyymm format
integer_date <- gridded_data[((i-1)*37+1),2]*100+gridded_data[((i-1)*37+1),1]
#convert to Date class
current_date <- ymd(integer_date ,truncated=1)
#extract named month
current_month <- month(current_date,label=T)
#extract year
current_year <- year(current_date)
#paste together named month and year to name data
names(monthly_data)[i] <- paste(current_month, current_year)
}
#for loop
for(i in 1:number_of_months){
#save selected month into dataset
current_data <- monthly_data[[i]]
#create latitude vector
latitudes <- rev(seq(from=-87.5, to=87.5, by=5))
#create latitude variable
current_data$latitude <- latitudes
#create longitude vector
longitudes <- seq(from=-177.5, to=177.5, by=5)
#make these values the column names
colnames(current_data)[1:72] <- longitudes
#gather all our temperature anomaly values into a single variable
current_data <- gather(current_data, key="longitude", value="TempAnom", -latitude)
#change class to numeric
current_data$longitude <- as.numeric(current_data$longitude)
#convert the -9999 to NA and convert to Celsius
current_data <- mutate(current_data,
TempAnom = ifelse(TempAnom==-9999, NA, TempAnom/100))
monthly_data[[i]] <- current_data
}
# To create graph for August 1916
library(maps)##
## Attaching package: 'maps'## The following object is masked from 'package:purrr':
##
## map#upload map into R
world <- map_data("world")
#set current month
current_month <- "Aug 1916"
#create variable
current_data <- monthly_data[[current_month]]
#create variable
current_name <- names(monthly_data[current_month])
#graph temperature anomoly in tile plot
g<-ggplot()+
geom_tile(data=current_data,
aes(x=longitude, y=latitude, fill=TempAnom),
width=5, height=5,alpha=.5)+
scale_fill_gradient2(low="blue",mid="white",high="red",midpoint=0)+
theme_void()+labs(title=current_name)
# project map onto tile plot
g <- g+geom_path(data = world, aes(x = long, y = lat, group = group))
g + coord_map("albers",parameters=c(0,0))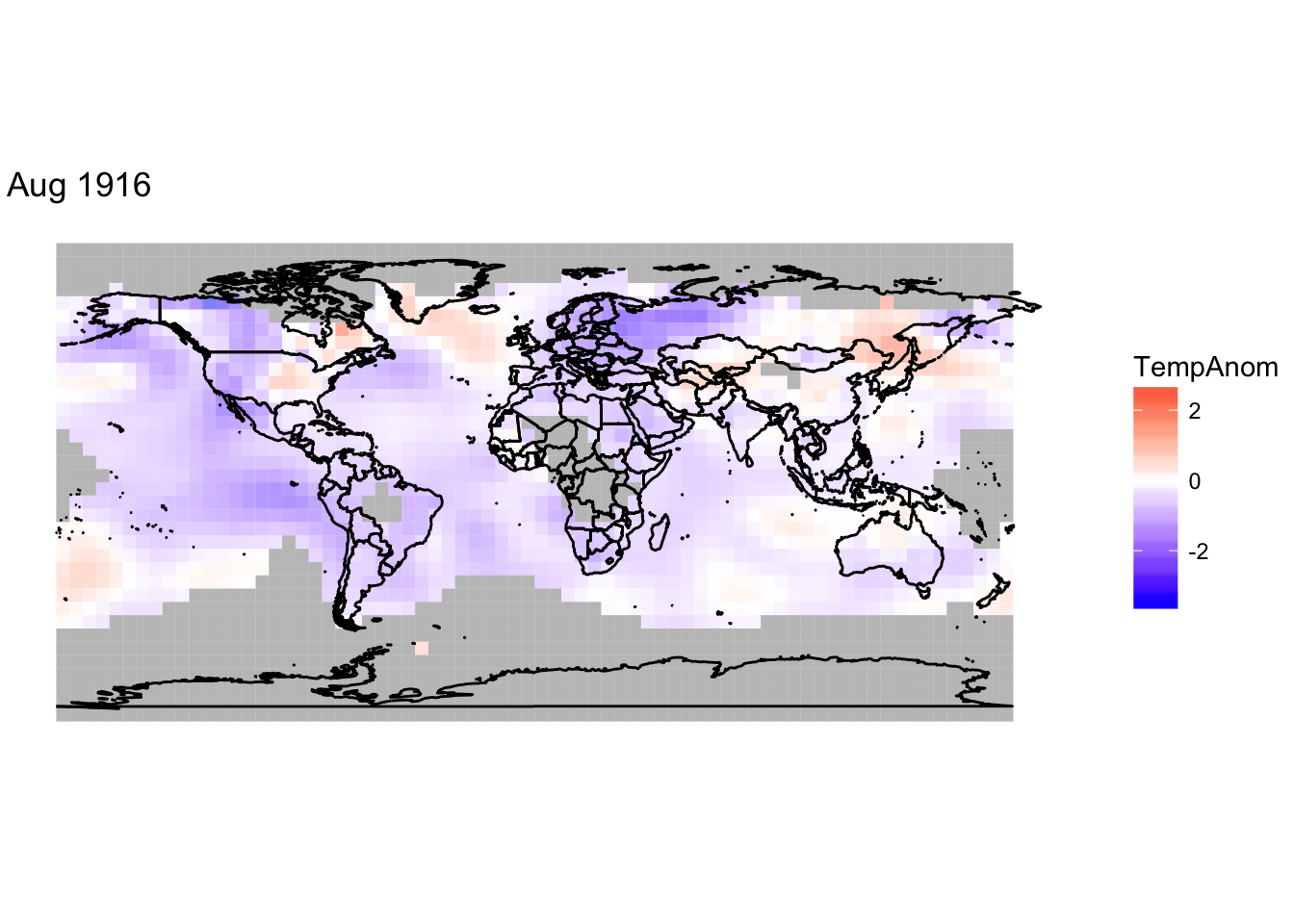
Exercise 3
#save manipulated data as new R file
save(monthly_temps, monthly_data, file="climate_data.RData")